Parasitic Plants
Molecular Phylogenetics of Scrophulariaceae/Orobanchaceae
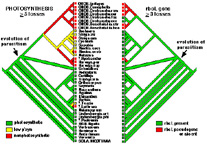
Our principal study group, the angiosperms formerly known as the parasitic Scrophulariaceae (figwort or snapdragon family) and Orobanchaceae (broomrape family), is the only lineage with a full range of parasites from photosynthetic plants with minimal parasitic ability and nonparasitic relatives, through to totally nonphotosynthetic members. It also includes the most economically-important parasitic weeds (Orobanche, Striga), which are responsible for huge amounts of crop loss worldwide. Species of parasitic Scrophulariaceae and Orobanchaceae disiplay a wide range of host preferences from broad (i.e., diverse dicot and monocot families) to very narrow (one species or subspecies). My lab has developed a molecular phylogenetic outline of parasitic evolution within the Scrophulariaceae using plastid gene sequences that are conserved and retained in all plants regardless of photosynthetic ability. These phylogenies have shown that parasitism evolved only once in the Lamiales, but the complete loss of photosynthesis within this group has occurred on numerous independent occasions. In addition to showing that parasitism originated just once within a highly paraphyletic "Scrophulariaceae" (Olmstead et al., 2001), our results have many additional implications for the origin, evolution, and classification of parasitic and nonparasitic Lamiales.
Molecular Evolution of Parasitic Plants
Molecular evolutionary studies of these parasitic plants has used the permanent loss of photosynthesis in several groups of parasites - as a means of 1) Understanding the molecular mechanisms of chloroplast DNA (cpDNA) evolution and 2) Revealing the potential contribution of plastid genes to nonphotosynthetic functions within the plastid. Detailed analysis of one parasitic Orobanchaceae, Epifagus virginiana, showed that this parasite has a plastid genome that is greatly reduced in size yet still functional. Comparative analysis has used the phylogeny as a powerful tool for resconstructing the history of these dramatic changes and investigating molecular evolution as a function of parasitic ability. Three plastid genes with different functions (rps2 - ribosomal protein; matK - intron splicing; and rbcL - photosynthesis) have now been analyzed in detail and provide important insights about the mechanisms that determine rates of sequence evolution, while allowing us to infer unexpected features about their molecular evolutions and function (Young and dePamphilis, 2005).
Future Studies
We are working to use the genomic and informatic methodologies we developed for the Floral Genome Project to study nuclear genomes in parasitic plants and to develop a detailed understanding of the evolutionary origin and function of genes that make the haustorium.
Interested in the possibility of research or study in the lab? Please email me.